Publications
Recent papers
(For a full list of publications and patents see below or go to Google Scholar, Pubmed)
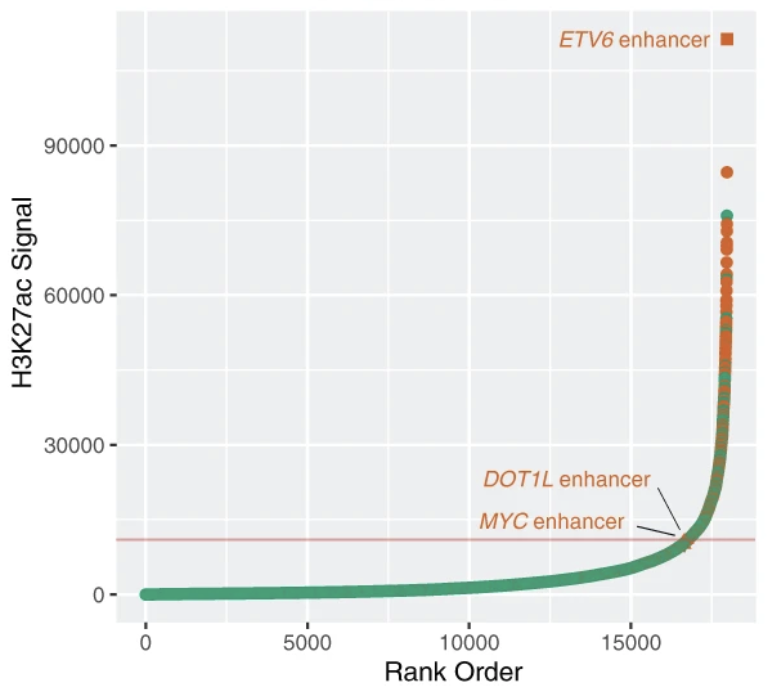
Comprehensive analysis of DNA methylation in primary AML samples with mutations in IDH1 and IDH2 demonstrated focal hypermethylation at regulatory enhancers, which may contribute to AML pathogenesis.
Wilson ER, Helton NM, Heath SE, Fulton RS, Payton JE, Welch JS, Walter MJ, Westervelt P, DiPersio JF, Link DC, Miller CA, Ley TJ, Spencer DH
Leukemia. 2022 Apr;36(4):935-945
Streamlined whole-genome sequencing for rapid, accurate, and acccessible genomic profiling of patients with AML and MDS.
Duncavage EJ, Schroeder MC, O’Laughlin M, Wilson R, MacMillan S, Bohannon A, Kruchowski S, Garza J, Du F, Hughes AEO, Robinson J, Hughes E, Heath SE, Baty JD, Neidich J, Christopher MJ, Jacoby MA, Uy GL, Fulton RS, Miller CA, Payton JE, Link DC, Walter MJ, Westervelt P, DiPersio JF, Ley TJ, Spencer DH.
N Engl J Med. 2021 Mar 11;384(10):924-935.
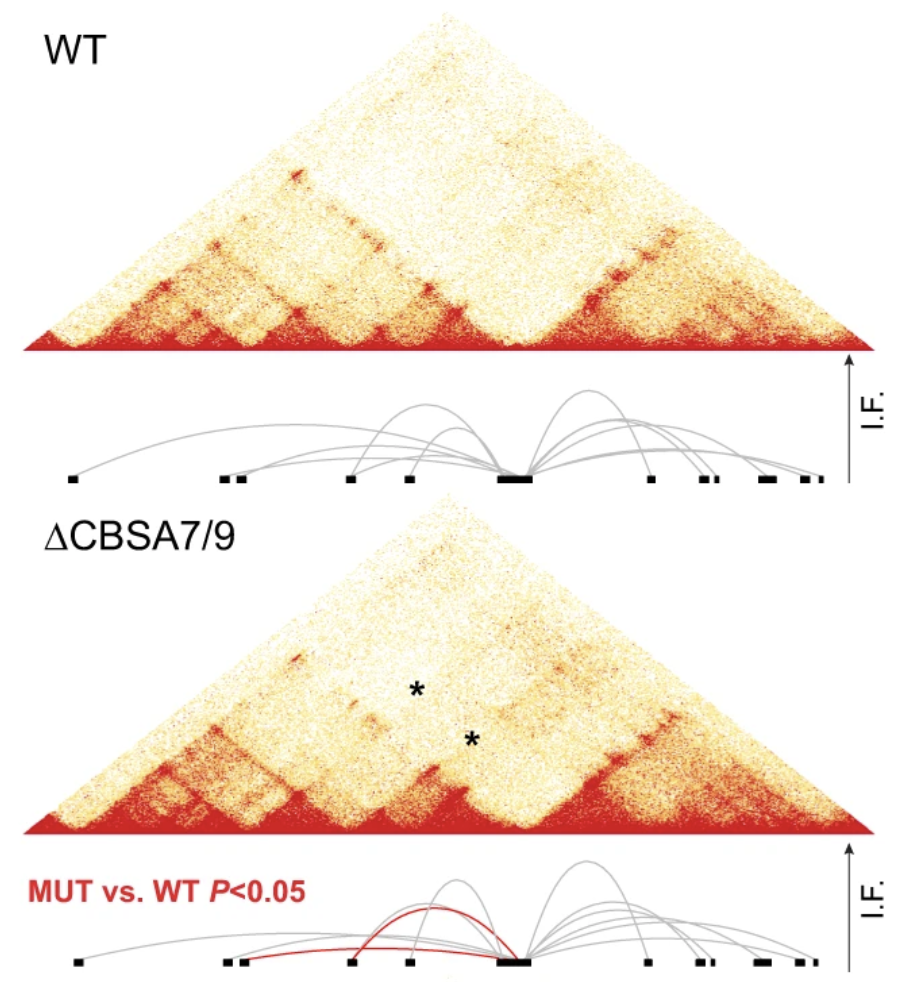
Analysis of epigenetic patterns and 3D genome architecture at the HOXA locus in primary AML samples with NPM1 mutations and genetic manipulation of CTCF binding sites in the NPM1-mutant OCI-AML3 cell line to define the role of CTCF binding in regulating HOXA genome interactions and gene expression.
Ghasemi R, Struthers H, Wilson ER, Spencer DH.
Leukemia. 2021 Feb;35(2):404-416.
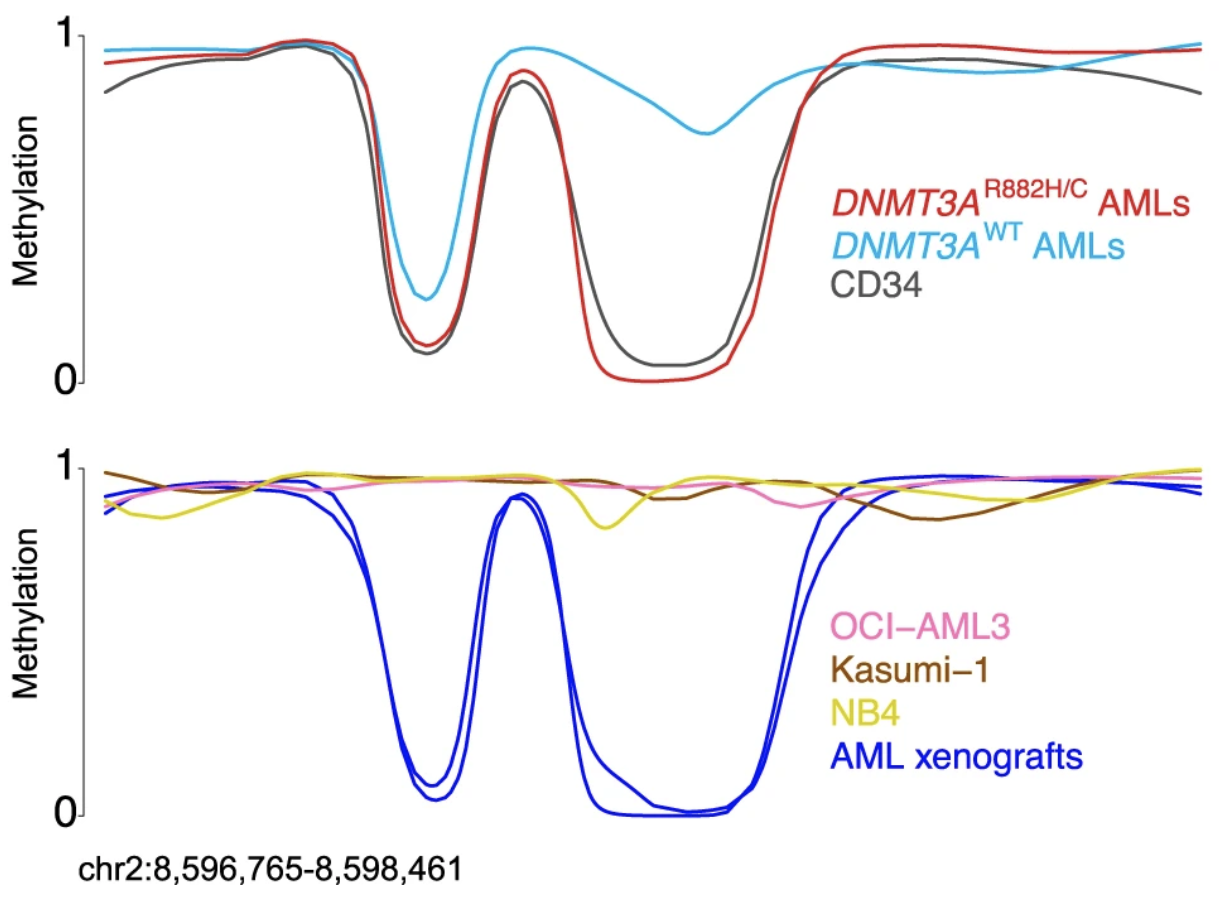
DNA methylation analysis of AML cell lines as appropriate models of AML-associated methylation phenotypes.
Chen D, Christopher M, Helton NM, Ferguson I, Ley TJ, Spencer DH.
Blood Cancer J. 2018 Apr 4;8(4):38.
Patents
Eric J Duncavage, David H Spencer
A streamlined
whole-genome sequencing approach for genetic profiling of cancer. (Pending) (2021)
More publications
Focal disruption of DNA methylation dynamics at enhancers in IDH-mutant AML cells
Wilson ER, Helton NM, Heath SE, Fulton RS, Payton JE, Welch JS, Walter MJ, Westervelt P, DiPersio JF, Link DC, Miller CA, Ley TJ, Spencer DH
Leukemia. 2022 Apr;36(4):935-945
Hematopoietic cell transplantation donor-derived memory-like NK cells functionally persist after transfer into patients with leukemia
Berrien-Elliott MM, Foltz JA, Russler-Germain DA, Neal CC, Tran J, Gang M, Wong P, Fisk B, Cubitt CC, Marin ND, Zhou AY, Jacobs MT, Foster M, Schappe T, McClain E, Kersting-Schadek S, Desai S, Pence P, Becker-Hapak M, Eisele J, Mosior M, Marsala L, Griffith OL, Griffith M, Khan SM, Spencer DH, DiPersio JF, Romee R, Uy GL, Abboud CN, Ghobadi A, Westervelt P, Stockerl-Goldstein K, Schroeder MA, Wan F, Lie WR, Soon-Shiong P, Petti AA, Cashen AF, Fehniger TA
Sci Transl Med. 2022 Feb 23;14(633):eabm1375
Genome Sequencing as an Alternative to Cytogenetic Analysis in Myeloid Cancers.
Duncavage EJ, Schroeder MC, O’Laughlin M, Wilson R, MacMillan S, Bohannon A, Kruchowski S, Garza J, Du F, Hughes AEO, Robinson J, Hughes E, Heath SE, Baty JD, Neidich J, Christopher MJ, Jacoby MA, Uy GL, Fulton RS, Miller CA, Payton JE, Link DC, Walter MJ, Westervelt P, DiPersio JF, Ley TJ, Spencer DH.
N Engl J Med. 2021 Mar 11;384(10):924-935.
Contribution of CTCF binding to transcriptional activity at the HOXA locus in NPM1-mutant AML cells.
Ghasemi R, Struthers H, Wilson ER, Spencer DH.
Leukemia. 2021 Feb;35(2):404-416.
Remethylation of Dnmt3a-/- hematopoietic cells is associated with partial correction of gene dysregulation and reduced myeloid skewing
Ketkar S, Verdoni AM, Smith AM, Bangert CV, Leight ER, Chen DY, Brune MK, Helton NM, Hoock M, George DR, Fronick C, Fulton RS, Ramakrishnan SM, Chang GS, Petti AA, Spencer DH, Miller CA, Ley TJ
Proc Natl Acad Sci U S A. 2020 Feb 11;117(6):3123-3134.
Inflammatory cytokines promote clonal hematopoiesis with specific mutations in ulcerative colitis patients
Zhang CRC, Nix D, Gregory M, Ciorba MA, Ostrander EL, Newberry RD, Spencer DH, Challen GA
Exp Hematol. 2019 Dec;80:36-41.e3.
Functional characterization of biallelic RTTN variants identified in an infant with microcephaly, simplified gyral pattern, pontocerebellar hypoplasia, and seizures
Wambach JA, Wegner DJ, Yang P, Shinawi M, Baldridge D, Betleja E, Shimony JS, Spencer D, Hackett BP, Andrews MV, Ferkol T, Dutcher SK, Mahjoub MR, Cole FS
Pediatr Res. 2018 Sep;84(3):435-441.
Sequencing of Tumor DNA to Guide Cancer Risk Assessment and Therapy
Spencer DH, Ley TJ
JAMA. 2018 Apr 10;319(14):1497-1498.
DNMT3AR882-associated hypomethylation patterns are maintained in primary AML xenografts, but not in the DNMT3AR882C OCI-AML3 leukemia cell line.
Chen D, Christopher M, Helton NM, Ferguson I, Ley TJ, Spencer DH.
Blood Cancer J. 2018 Apr 4;8(4):38.
Expression profiling of snoRNAs in normal hematopoiesis and AML
Warner WA, Spencer DH, Trissal M, White BS, Helton N, Ley TJ, Link DC
Blood Adv. 2018 Jan 23;2(2):151-163.
Comprehensive discovery of noncoding RNAs in acute myeloid leukemia cell transcriptomes
Zhang J, Griffith M, Miller CA, Griffith OL, Spencer DH, Walker JR, Magrini V, McGrath SD, Ly A, Helton NM, Trissal M, Link DC, Dang HX, Larson DE, Kulkarni S, Cordes MG, Fronick CC, Fulton RS, Klco JM, Mardis ER, Ley TJ, Wilson RK, Maher CA
Exp Hematol. 2017 Nov;55:19-33.
CpG Island Hypermethylation Mediated by DNMT3A Is a Consequence of AML Progression
Spencer DH, Russler-Germain DA, Ketkar S, Helton NM, Lamprecht TL, Fulton RS, Fronick CC, O’Laughlin M, Heath SE, Shinawi M, Westervelt P, Payton JE, Wartman LD, Welch JS, Wilson RK, Walter MJ, Link DC, DiPersio JF, Ley TJ
Cell. 2017 Feb 23;168(5):801-816.e13.
CIViC is a community knowledgebase for expert crowdsourcing the clinical interpretation of variants in cancer
Griffith M, Spies NC, Krysiak K, McMichael JF, Coffman AC, Danos AM, Ainscough BJ, Ramirez CA, Rieke DT, Kujan L, Barnell EK, Wagner AH, Skidmore ZL, Wollam A, Liu CJ, Jones MR, Bilski RL, Lesurf R, Feng YY, Shah NM, Bonakdar M, Trani L, Matlock M, Ramu A, Campbell KM, Spies GC, Graubert AP, Gangavarapu K, Eldred JM, Larson DE, Walker JR, Good BM, Wu C, Su AI, Dienstmann R, Margolin AA, Tamborero D, Lopez-Bigas N, Jones SJ, Bose R, Spencer DH, Wartman LD, Wilson RK, Mardis ER, Griffith OL
Nat Genet. 2017 Jan 31;49(2):170-174.
Occult Specimen Contamination in Routine Clinical Next-Generation Sequencing Testing
Sehn JK, Spencer DH, Pfeifer JD, Bredemeyer AJ, Cottrell CE, Abel HJ, Duncavage EJ
Am J Clin Pathol. 2015 Oct;144(4):667-74.
Association Between Mutation Clearance After Induction Therapy and Outcomes in Acute Myeloid Leukemia
Klco JM, Miller CA, Griffith M, Petti A, Spencer DH, Ketkar-Kulkarni S, Wartman LD, Christopher M, Lamprecht TL, Helton NM, Duncavage EJ, Payton JE, Baty J, Heath SE, Griffith OL, Shen D, Hundal J, Chang GS, Fulton R, O’Laughlin M, Fronick C, Magrini V, Demeter RT, Larson DE, Kulkarni S, Ozenberger BA, Welch JS, Walter MJ, Graubert TA, Westervelt P, Radich JP, Link DC, Mardis ER, DiPersio JF, Wilson RK, Ley TJ
JAMA. 2015 Aug 25;314(8):811-22.
Epigenomic analysis of the HOX gene loci reveals mechanisms that may control canonical expression patterns in AML and normal hematopoietic cells
Spencer DH, Young MA, Lamprecht TL, Helton NM, Fulton R, O’Laughlin M, Fronick C, Magrini V, Demeter RT, Miller CA, Klco JM, Wilson RK, Ley TJ
Leukemia. 2015 Jun;29(6):1279-89.
Clinical next-generation sequencing in patients with non-small cell lung cancer
Hagemann IS, Devarakonda S, Lockwood CM, Spencer DH, Guebert K, Bredemeyer AJ, Al-Kateb H, Nguyen TT, Duncavage EJ, Cottrell CE, Kulkarni S, Nagarajan R, Seibert K, Baggstrom M, Waqar SN, Pfeifer JD, Morgensztern D, Govindan R
Cancer. 2015 Feb 15;121(4):631-9.
The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers
Russler-Germain DA, Spencer DH, Young MA, Lamprecht TL, Miller CA, Fulton R, Meyer MR, Erdmann-Gilmore P, Townsend RR, Wilson RK, Ley TJ
Cancer Cell. 2014 Apr 14;25(4):442-54.
Functional heterogeneity of genetically defined subclones in acute myeloid leukemia
Klco JM, Spencer DH, Miller CA, Griffith M, Lamprecht TL, O’Laughlin M, Fronick C, Magrini V, Demeter RT, Fulton RS, Eades WC, Link DC, Graubert TA, Walter MJ, Mardis ER, Dipersio JF, Wilson RK, Ley TJ
Cancer Cell. 2014 Mar 17;25(3):379-92.
Performance of common analysis methods for detecting low-frequency single nucleotide variants in targeted next-generation sequence data
Spencer DH, Tyagi M, Vallania F, Bredemeyer AJ, Pfeifer JD, Mitra RD, Duncavage EJ
J Mol Diagn. 2014 Jan;16(1):75-88.
Validation of a next-generation sequencing assay for clinical molecular oncology
Cottrell CE, Al-Kateb H, Bredemeyer AJ, Duncavage EJ, Spencer DH, Abel HJ, Lockwood CM, Hagemann IS, O’Guin SM, Burcea LC, Sawyer CS, Oschwald DM, Stratman JL, Sher DA, Johnson MR, Brown JT, Cliften PF, George B, McIntosh LD, Shrivastava S, Nguyen TT, Payton JE, Watson MA, Crosby SD, Head RD, Mitra RD, Nagarajan R, Kulkarni S, Seibert K, Virgin HW 4th, Milbrandt J, Pfeifer JD
J Mol Diagn. 2014 Jan;16(1):89-105.
Comparison of clinical targeted next-generation sequence data from formalin-fixed and fresh-frozen tissue specimens
Spencer DH, Sehn JK, Abel HJ, Watson MA, Pfeifer JD, Duncavage EJ
J Mol Diagn. 2013 Sep;15(5):623-33.
Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia
Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, Robertson A, Hoadley K, Triche TJ Jr, Laird PW, Baty JD, Fulton LL, Fulton R, Heath SE, Kalicki-Veizer J, Kandoth C, Klco JM, Koboldt DC, Kanchi KL, Kulkarni S, Lamprecht TL, Larson DE, Lin L, Lu C, McLellan MD, McMichael JF, Payton J, Schmidt H, Spencer DH, Tomasson MH, Wallis JW, Wartman LD, Watson MA, Welch J, Wendl MC, Ally A, Balasundaram M, Birol I, Butterfield Y, Chiu R, Chu A, Chuah E, Chun HJ, Corbett R, Dhalla N, Guin R, He A, Hirst C, Hirst M, Holt RA, Jones S, Karsan A, Lee D, Li HI, Marra MA, Mayo M, Moore RA, Mungall K, Parker J, Pleasance E, Plettner P, Schein J, Stoll D, Swanson L, Tam A, Thiessen N, Varhol R, Wye N, Zhao Y, Gabriel S, Getz G, Sougnez C, Zou L, Leiserson MD, Vandin F, Wu HT, Applebaum F, Baylin SB, Akbani R, Broom BM, Chen K, Motter TC, Nguyen K, Weinstein JN, Zhang N, Ferguson ML, Adams C, Black A, Bowen J, Gastier-Foster J, Grossman T, Lichtenberg T, Wise L, Davidsen T, Demchok JA, Shaw KR, Sheth M, Sofia HJ, Yang L, Downing JR, Eley G
N Engl J Med. 2013 May 30;368(22):2059-74.
Genomic impact of transient low-dose decitabine treatment on primary AML cells
Klco JM, Spencer DH, Lamprecht TL, Sarkaria SM, Wylie T, Magrini V, Hundal J, Walker J, Varghese N, Erdmann-Gilmore P, Lichti CF, Meyer MR, Townsend RR, Wilson RK, Mardis ER, Ley TJ
Blood. 2013 Feb 28;121(9):1633-43.
Detection of FLT3 internal tandem duplication in targeted, short-read-length, next-generation sequencing data
Spencer DH, Abel HJ, Lockwood CM, Payton JE, Szankasi P, Kelley TW, Kulkarni S, Pfeifer JD, Duncavage EJ
J Mol Diagn. 2013 Jan;15(1):81-93.
Direct-to-consumer genetic testing: reliable or risky?
Spencer DH, Lockwood C, Topol E, Evans JP, Green RC, Mansfield E, Tezak Z
Clin Chem. 2011 Dec;57(12):1641-4.
Validation and implementation of the GeneXpert MRSA/SA blood culture assay in a pediatric setting
Spencer DH, Sellenriek P, Burnham CA
Am J Clin Pathol. 2011 Nov;136(5):690-4.
Red cell transfusion decreases hemoglobin A1c in patients with diabetes
Spencer DH, Grossman BJ, Scott MG
Clin Chem. 2011 Feb;57(2):344-6.
IFRD1 is a candidate gene for SMNA on chromosome 7q22-q23
Brkanac Z, Spencer D, Shendure J, Robertson PD, Matsushita M, Vu T, Bird TD, Olson MV, Raskind WH
Am J Hum Genet. 2009 May;84(5):692-7.
Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients
D’Argenio DA, Wu M, Hoffman LR, Kulasekara HD, Déziel E, Smith EE, Nguyen H, Ernst RK, Larson Freeman TJ, Spencer DH, Brittnacher M, Hayden HS, Selgrade S, Klausen M, Goodlett DR, Burns JL, Ramsey BW, Miller SI
Mol Microbiol. 2007 Apr;64(2):512-33.
Detecting disease-causing mutations in the human genome by haplotype matching
Spencer DH, Bubb KL, Olson MV
Am J Hum Genet. 2006 Nov;79(5):958-64.
Evidence for diversifying selection at the pyoverdine locus of Pseudomonas aeruginosa
Smith EE, Sims EH, Spencer DH, Kaul R, Olson MV
J Bacteriol. 2005 Mar;187(6):2138-47.
Genome mosaicism is conserved but not unique in Pseudomonas aeruginosa isolates from the airways of young children with cystic fibrosis
Ernst RK, D’Argenio DA, Ichikawa JK, Bangera MG, Selgrade S, Burns JL, Hiatt P, McCoy K, Brittnacher M, Kas A, Spencer DH, Olson MV, Ramsey BW, Lory S, Miller SI
Environ Microbiol. 2003 Dec;5(12):1341-9.
Whole-genome sequence variation among multiple isolates of Pseudomonas aeruginosa
Spencer DH, Kas A, Smith EE, Raymond CK, Sims EH, Hastings M, Burns JL, Kaul R, Olson MV
J Bacteriol. 2003 Feb;185(4):1316-25.
Genetic variation at the O-antigen biosynthetic locus in Pseudomonas aeruginosa
Raymond CK, Sims EH, Kas A, Spencer DH, Kutyavin TV, Ivey RG, Zhou Y, Kaul R, Clendenning JB, Olson MV
J Bacteriol. 2002 Jul;184(13):3614-22.
Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen
Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock RE, Lory S, Olson MV
Nature. 2000 Aug 31;406(6799):959-64.