Clinical Genomics
David Spencer is a board-certified Molecular Pathologist and Medical Director of the McDonnell Genome Institute’s CLIA Licensed Environment (MGI-CLE), a CLIA-licensed, CAP-accredited clinical sequencing laboratory. He and his colleagues in the MGI-CLE and the Department of Pathology and Immunology have developed and implemented multiple advanced clinical sequencing assays for both translational research and patient care.


In his role as Medical Director, David oversees the design, development, and implementation of all NGS assays in the MGI Clinical sequencing lab, including wet lab and bioinformatics. He reviews all documentation and quality control data to maintain compliance with CLIA regulations and CAP checklist requirements, and serves as a consultant to investigators and clinicians who want to incorporate NGS testing into their research and/or clinical practice.
Current CLIA-compliant NGS assays
 ChromoSeq: Streamlined
whole-genome sequencing for cancers. Dr. Spencer and his colleagues Dr. Molly Schroeder and
Dr. Eric Duncavage developed this assay to provide comprehensive
genomic profiling of acute myeloid leukemia (AML) and
myelodysplastic syndromes (MDS) and published it in the New England
Journal of
Medicine in
March 2021. This publication was the first to demonstrate that
genome sequencing can be used in a clinical setting for fast,
accurate, and accessible genetic testing of tumors. It is currently
being performed on all new AML patients at Barnes-Jewish Hospital.
ChromoSeq: Streamlined
whole-genome sequencing for cancers. Dr. Spencer and his colleagues Dr. Molly Schroeder and
Dr. Eric Duncavage developed this assay to provide comprehensive
genomic profiling of acute myeloid leukemia (AML) and
myelodysplastic syndromes (MDS) and published it in the New England
Journal of
Medicine in
March 2021. This publication was the first to demonstrate that
genome sequencing can be used in a clinical setting for fast,
accurate, and accessible genetic testing of tumors. It is currently
being performed on all new AML patients at Barnes-Jewish Hospital.
MyeloSeq: Error-corrected targeted sequencing for diagnosis and molecular monitoring of patients with myeloid malignancies. This assay uses Agilent’s HaloplexHS unique molecular identifier (UMI)-based amplicon system to sequence 48 genes that are recurrently mutated in AML, MDS, and MPN. High coverage sequencing, error-correction, and custom analysis is performed using approaches developed by Eric Duncavage, Haley Abel, and David Spencer and is coded in WDL and executed via Cromwell.
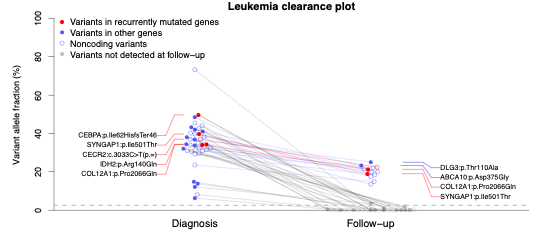 AML Clearance Exome Sequencing: This assay uses deep exome
sequencing to identify dozens of mutations in the leukemia cells of
AML patients at diagnosis, and then test whether these
patient-specific mutations are present in exome sequencing data from
the patient’s bone marrow cells
after they receive therapy. Previous studies have shown that the
presence of persistent mutations is associated with worse outcomes,
even when patients have no detectable leukemia cells in their bone
marrow. This assay is the core technology underlying a prospective
clinical trial to determine whether molecular monitoring using this
approach can improve outcomes for AML patients (NCT02756962).
AML Clearance Exome Sequencing: This assay uses deep exome
sequencing to identify dozens of mutations in the leukemia cells of
AML patients at diagnosis, and then test whether these
patient-specific mutations are present in exome sequencing data from
the patient’s bone marrow cells
after they receive therapy. Previous studies have shown that the
presence of persistent mutations is associated with worse outcomes,
even when patients have no detectable leukemia cells in their bone
marrow. This assay is the core technology underlying a prospective
clinical trial to determine whether molecular monitoring using this
approach can improve outcomes for AML patients (NCT02756962).
Tumor/Normal Exome Sequencing: Paired exome sequencing of tumor DNA and normal DNA from the same patient to identify somatic mutations. Analysis uses a custom ensemble pipeline encoded in CWL and executed with the Cromwell engine. This assay can be performed on any tumor type or specimen and is used to identify somatic mutations for translational studies using somatic mutations to manufacture patient-specific tumor vaccines.
Exome Sequencing (technical sequencing only): This assay uses hybrid-capture with a custom version of the IDT xGen Exome probeset for exome sequencing. This is a technical sequencing assay, and so analysis is performed for QC only. Variant analysis and all clinical interpretation is performed in the Laboratory and Genomic Medicine Division in the Department of Pathology and Immunology.
Comprehensive Cancer Gene Panel (technical sequencing only): High-coverage targeted sequencing of cancer-related genes. This assay also uses hybrid-capture with a custom IDT xGen probeset and is a technical sequencing only assay. Variant analysis and all clinical interpretation is performed in the Laboratory and Genomic Medicine Division in the Department of Pathology and Immunology.
Clinical sequencing assays in the MGI-CLE are available for patient care through either the AMP Core Labsin the Anatomic and Molecular Pathology Divsion or the Laboratory and Genomic Medicine Division in the Department of Pathology and Immunology.
David is Technical Director of the AMP Core Labs and works with faculty and staff in the AMP division to manage the ‘NGS distributed testing’ workflow for cancer testing testing performed in the MGI-CLE lab.